Abstract
Background:
Follicular lymphoma (FL) is the most common indolent non-Hodgkin's lymphoma. FL is often responsive to standard treatment but remains incurable with conventional therapies, and may transform into more aggressive lymphoma types. The mutational processes that leads to lymphomagenesis and progression also produce tumor-specific mutant antigens (TSMAs) that can be targeted by the immune system to control malignancies. Personalized cancer vaccines designed for these TSMAs represent a promising new strategy for treatment of FL. However, the feasibility of this approach and precisely how to optimize effective vaccine design, informed by next-generation sequencing data, is not fully understood. We hypothesize that (tumor/normal) whole exome sequencing (WES) and (tumor) RNA sequencing (RNA-Seq) can be used to predict patient HLA typing and neoepitopes to engineer personalized cancer vaccines for FL.
Methods:
Using genomic DNA and RNA from FL patient biopsy specimens, we performed WES and RNA-Seq on fresh frozen primary or relapsed tumor samples from 33 FL patients with matched normal skin. This sequencing achieved >20x coverage for >75% of the targeted regions with a mean coverage of 76x. From this analysis, we identified an average of 145 somatic mutations per patient (range: 15-419). RNA-seq data allowed us to identify the expressed neoantigens. To determine the patient's HLA-A, HLA-B and HLA-C typing we used both OptiType and ATHLATES, achieving concordance between these algorithms of 80%, 94.29% and 90%, respectively. We used the Variant Effect Predictor (VEP) software for annotation of protein-coding effect of variants. Epitope predictions were performed for each patient's mutation(s) against all 6 of their HLA class I alleles using pVAC-Seq, version 4.0.8 (Hundal et al. Genome Med. 2016). Mutations were condensed to the single best neoepitope candidate. All patient-predicted peptides for a single variant were filtered to identify high quality candidates using the following criteria: (1) HLA binding affinity IC50 ≤ 500 nmol/L; (2) normal/tumor DNA/RNA coverage > 10; (3) normal DNA variant allele frequency (VAF) < 2; (4) tumor DNA/RNA VAF > 10; and (5) gene and transcript expression > 0.
Results:
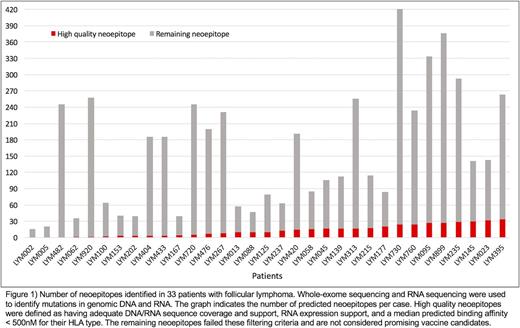
High quality neoepitopes were predicted for all but three patients (90.9%). Five or more neoepitope candidates were identified for 22 (66.7%) patients with a mean of 12 predicted peptides per patient (range: 0-33). Overall, 80% (342) of the total predicted patient peptides arose from missense mutations and the remaining 20% (81) from indels. Predicted neoepitopes were identified in genes known to be recurrently mutated in lymphoma, including CREBBP, KMT2D, TNFRSF14, BCL2, BCL7A, EZH2, ARID1A/B, ATP6V1B2, CD22, CD79B, CXCR4, HIST1H1/H2 family members, HVCN1, IRF8, MEF2B, POU2F2, POU2AF1, PIM1, RRAGC, TP53, ZNF608, and IGLL5.
Conclusion:
Peptides suitable for cancer vaccine generation were identified for 90.9% (30 of 33 patients) of our cohort via next-generation sequencing, pVAC-Seq and filtering techniques. These neoepitopes include those which arose from genes known to be recurrently mutated in follicular lymphoma. Collectively, these findings suggest that most follicular lymphoma patients will be candidates for clinical trials that incorporate personalized cancer vaccines.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal